Premise
How can a deep learning image classification tool be used to identify invasive plant species?
Synopsis
Biological invasions, where a non-native species is introduced to a new environment where it proliferates and dominates, i.e., becomes invasive, drastically altering the functions of that ecosystem, are one of the biggest drivers of biodiversity loss globally. Once an invasive species becomes established in a new environment, it is crucial to detect it early to minimise the potential harm it can cause. Traditionally, the detection of invasive species had to be carried out by teams of experts who would systematically sample predetermined areas. These methods are costly, inefficient and unscalable. Increasingly, deep learning techniques are being used to detect invasive species with much greater efficiency. Here, I try to build on these innovations by developing an image classification tool that identifies invasive species from images taken by members of the general public.
Substantiation
‘Biodiversity’ broadly refers to the number, abundance, composition, spatial distribution, and interactions of the approximately 9 million different types of organisms that live on Earth (Díaz et al., 2006). The survival of humanity is dependent on biodiversity, as biodiversity influences the ecosystem services that purify air and water, regulate the climate, control pests and diseases etc., all of which make life possible and worth living (Cardinale et al., 2012). Despite this, species are being lost from ecosystems at an alarming rate. (Heywood & Watson, 1995). One of the biggest threats to biodiversity are biological invasions, which occur when a species is transported to a new environment where they can proliferate, spread, and persist, (Elton, 1958) altering the functions of that ecosystem (Mack et al., 2000). It is difficult and costly to control a species once it becomes established (Sanders et al., 2019). Early detection of an invasive species is crucial to minimise the potential consequences (Simberloff, 1997). Unfortunately, the monitoring of invasive species is usually insufficient (Mack et al., 2000), as specialists are required on-site to sample a designated area, which is inefficient and expensive (Ashqar & Abu-Naser, 2019).
This is where deep learning solutions offer a lot of promise. These techniques are increasingly being employed to identify, monitor and track invasive species, allowing for greater areas to be covered at fractions of the cost and time, with similar levels of accuracy (Qian et al., 2020). Several studies have developed image recognition and computer vision tools to identify invasive species in various settings (Ashqar & Abu-Nasser, 2019; Martínez-González et al., 2021; Klein et al., 2015). In this study, I seek to build on these innovations to develop an image classification tool to identify invasive plant species.
Preparation
Following a thorough literature review of relevant studies, I determined that it would not be feasible to develop my own classifier within the short timeframe as there is a steep learning curve. Instead, I will use a tool called Lobe, which makes the process of training a machine learning model easier and faster.
Finding a suitable dataset of invasive plant species was difficult as most online were either too small or under private license I was also unable to retrieve a large enough dataset through the Bing API. In the end I opted to use a Kaggle dataset that contained ~3800 images of an invasive species of hydrangea and other non-invasive plants. My goal, therefore, would be to build a model that would be able to classify images in the dataset as either invasive or non-invasive, providing a proof-of-concept for further studies in this area.
First Iteration
After downloading the dataset from Kaggle, I found that the images were split into test/train folders, images of both the invasive and non-invasive species were in one folder, the labels were in a separate file, and there were no labels for the test images, making them unusable. I wrote a python script that matched the training images to their label and moved them into a relevant folder for invasive/non-invasive species. There are 1448 images of the invasive species but only 847 images of non-invasive species. Lobe advises to use similar numbers of images for each class, so I will only use 847 of the images of invasive species. It is best practice to use larger datasets to better train deep learning algorithms which makes it unfortunate that I must limit the dataset for now. After training the Lobe model with the dataset, it performed with 92% accuracy.


The target audience for the model is people in the 18-30 age group because they are the youngest adult demographic and so are the most likely to experience the harshest effects of biodiversity loss. I interviewed 5 individuals, discussing the model and its purpose, to determine their motivations for using the model, and ways to improve the model to make it more enticing to use. The feedback was largely positive, but users felt that they would not be motivated to use the model long-term. A suggestion was made to widen the scope of the model to be able to identify plant species rather than simply classify them as invasive/non-invasive. This would attract a wider userbase such as gardeners or house plant buyers etc. This feedback provided me with a clear objective for the next iteration; add images of a broader range of plants to the dataset and more classes to the model to identify the plant species.
Second Iteration
I found several datasets from different sources that included images of different flowers. After reviewing, I selected 5 flower types to include in the model. After combining datasets, some data preparation and cleaning, I had a dataset that was ready to use.
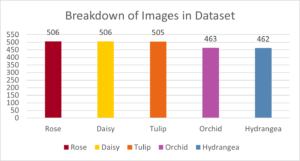
The model performed well with a classification accuracy of 95%. A sample of the correct and incorrect classifications can be seen below:
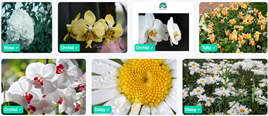

This is a positive result and users were pleased that the model now has more functionality and use cases. However, the model is trained on images that were sourced online, often taken professionally. The goal of this model is to allow anybody take a photo on their phone to identify a plant species. To test if the model works as well with user-taken photos, I enlisted the same 5 users to take photos of these types of flowers that they could find in their vicinity to test the model. I found that different users take very different photos, close-up, far back, portrait, landscape, and some included their hands holding the flower in frame.




The model performed with 70% accuracy in these tests, dealing with the inclusion of hands well, but struggling with photos taken from far away where the flower is smaller in the frame. To improve the model’s performance, it is necessary to include more photos in the training data that cover the many ways a user could take a photo.
Sample Correct Predictions



Sample Incorrect Predictions



Third Iteration
For this iteration, I sought to gather hundreds of user-taken images of the designated flowers to add to the dataset to improve performance. Unfortunately, this proved more difficult than imagined as I was unable to find many flowers in my area to create a large enough dataset. What photos I was able to gather, however, increased the model’s accuracy to 99%.
To further improve the model in this iteration, I added more flower classes, to give the model wider usability.
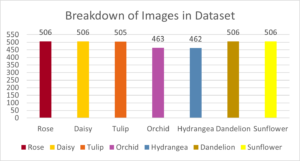
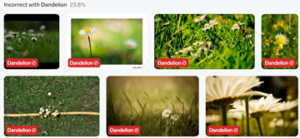
With the added data, the model performed with 90% accuracy, a considerable decrease. The model particularly struggled in identifying daisies, having only 68% accuracy. The best way to improve this in future iterations would be to simply add more training data.
Conclusion
Clearly, the model performs well as an image classifier. The challenge, without being able to finetune the code, comes down to having large amounts of data that are representative of the model’s intended use.
Reflection
This study was designed to be a starting point and proof of concept for my graduation assignment (GA). It has been successful in showing that deep learning can be successfully applied to the problem of identifying invasive plant species. I have learned much about how image classification models are trained, and it has inspired me to delve deeper into this technology and acquire the skills necessary to develop my own model. This study has validated much of the dataset I hoped to use for my GA and through the iterations, I have learned valuable lessons about the size and variety of the data needed to train a successful model. User testing also provided valuable insights into how and why users would use such an application. I learned that my initial idea of an invasive species identifier did not have a strong enough pull factor to get people to use the app in the long-term, so for my GA I will shift the focus to plant/flower identification. This makes the app more appealing to a wider range of users including gardeners, photographers, and homeowners, while I am still able to achieve the goal of identifying invasive species by matching the images taken to a list of invasive species of concern. I now have a clear idea of the direction I want to take my GA.
References
Ashqar, B. A., & Abu-Naser, S. S. (2019). Identifying images of invasive hydrangea using pre-trained deep convolutional neural networks. International Journal of Academic Engineering Research (IJAER), 3(3), 28-36.
Cardinale, B. J., Duffy, J. E., Gonzalez, A., Hooper, D. U., Perrings, C., Venail, P., … & Naeem, S. (2012). Biodiversity loss and its impact on humanity. Nature, 486(7401), 59-67.
Díaz, S., Fargione, J., Chapin III, F. S., & Tilman, D. (2006). Biodiversity loss threatens human well-being. PLoS biology, 4(8), e277.
Elton, C. S. (1958). The ecology of invasions by animals and plants. Methuen.
Heywood, V. H., & Watson, R. T. (1995). Global biodiversity assessment (Vol. 1140). Cambridge: Cambridge university press.
Klein, D. J., McKown, M. W., & Tershy, B. R. (2015, May). Deep learning for large scale biodiversity monitoring. In Bloomberg Data for Good Exchange Conference.
Mack, R. N., Simberloff, D., Mark Lonsdale, W., Evans, H., Clout, M., & Bazzaz, F. A. (2000). Biotic invasions: causes, epidemiology, global consequences, and control. Ecological applications, 10(3), 689-710.
Martínez-González, Á.T., Ramírez-Rivera, V.M., Caballero-Vázquez, J.A. et al. Deep learning algorithm as a strategy for detection an invasive species in uncontrolled environment. Rev Fish Biol Fisheries 31, 909–922 (2021).
Sanders, M.E., R.J.H.G. Henkens & D.M.E. Slijkerman (2019). Convention on Biological Diversity; Sixth National Report of the Kingdom of the Netherlands. Wageningen, the Statutory Research Tasks Unit for Nature & the Environment (WOT Natuur & Milieu). WOt-technical report 156.
Simberloff, D. 1997. Eradication. Pages 221–228 in D. Simberloff, D. C. Schmitz, and T. C. Brown, editors. Strangers in paradise.
Qian, W., Huang, Y., Liu, Q., Fan, W., Sun, Z., Dong, H., … & Qiao, X. (2020). UAV and a deep convolutional neural network for monitoring invasive alien plants in the wild. Computers and Electronics in Agriculture, 174, 105519.